Different Approaches to The Development of Nanobodies
Nanobodies and Their Unique Advantages
A new type of antibody naturally found in camelid, such as alpacas, differs in structure from traditional antibodies. This antibody consists of only two heavy chains, called heavy-chain antibody (HCAb).It lacks light chain and CH1 structures in heavy chain but retains complete antigen-binding activity. This heavy-chain variable region, which can specifically bind antigen, is called single-domain antibodies (sdAbs), or Nanobody, or VHH, which is the smallest unit of the antigen binding fragment.
Compared to conventional antibodies, nanobodies have a small molecular weight and simple structure. Based on the advantage of small molecular weight, nanobodies furthermore have several features that make them show great potential for new drug discovery. Nanobodies exhibit high affinity, high specificity, strong penetration and ease of modification and expression, and the complete antibody sequence was easy to obtain. Meanwhile, nanobodies can be produced in high quality and stable by in vitro recombinant expression, effectively avoiding the problem of batch-to-batch variation of traditional antibodies.
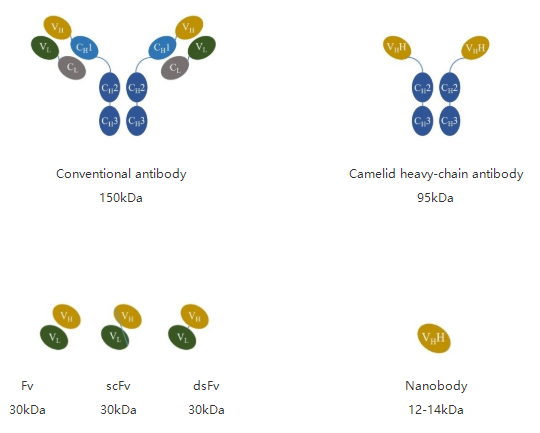
Figure 1. Different antibody types and molecular weights
Screening and Preparation of Nanobodies
Nanobodies are now commonly obtained through alpacas to obtain antibody genes, which are then screened by library display techniques to obtain the most suitable antibody sequences.
Based on phage display, the construction of a nanobody gene library is a technical pathway to obtain nanobody sequences. Phage libraries are mainly divided into natural library, immune library and synthesis library. Natural library constructs antibody genes from alpaca peripheral blood lymphocytes, bone marrow, spleen cells and tissues, and then clone these fragments directly into expression vectors for expression and display. Although natural library can avoid the time-consuming and laborious immunization process, they require larger library capacity and the affinity of the obtained nanobodies are most likely lower than that of immune libraries.
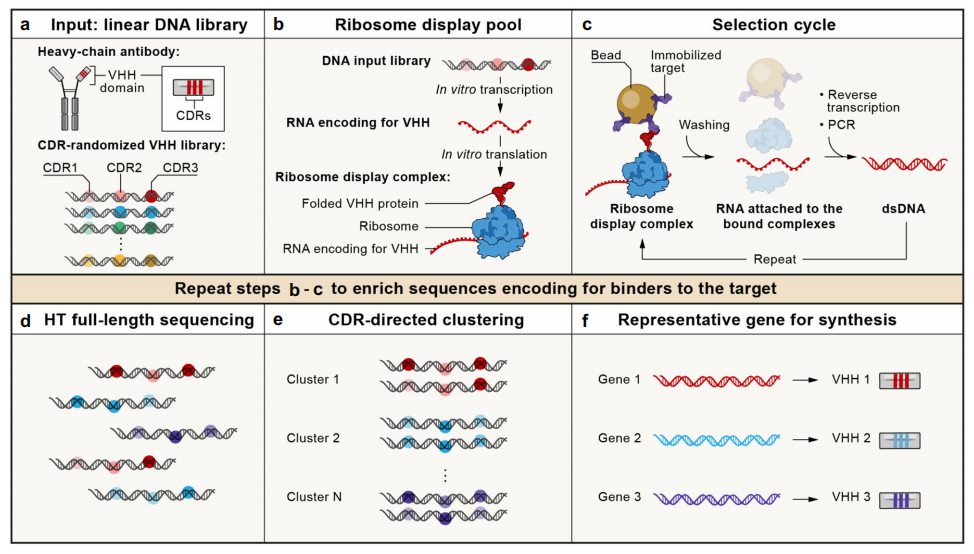
Figure 2. A cell-free nanobody engineering platform.
(Chen, Gentili, Hacohen & Regev, 2021)
To construct an immune library, albaca were immunized with antigens to extract lymphocytes containing mature B cells. The RNA of the B cells is extracted and inverted into a cDNA library, and the specific gene fragment is obtained by PCR amplification again in the cDNA library using specially designed primers. At last, subsequently, the nanobodies are displayed on the phage surface, and after the positive clones are sorted by liquid-phase or solid-phase, the diverse antibody sequences can be obtained by sequencing.
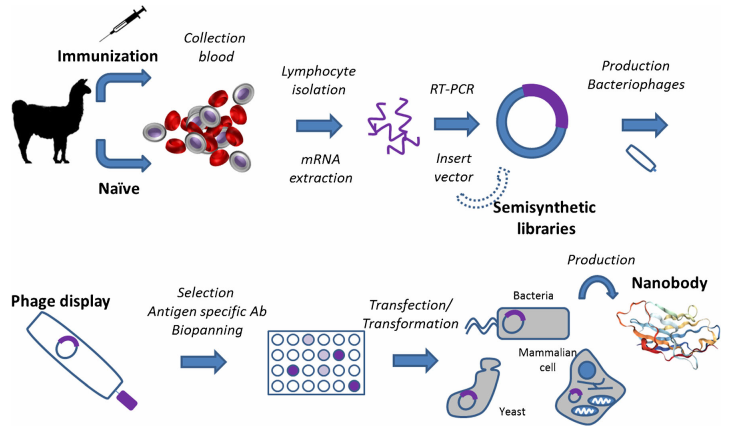
Figure 3. Nanobody production scheme using a phage display library.
(Salvador, Vilaplana & Marco, 2019)
The synthesis library is based on antibody sequences with good expression characters and strong stability as the framework to improve the expression level of antibody in the library and reduce the toxicity of the expressed products to the host cells. By increasing the proportion of functional antibody molecules in the antibody library, the real capacity of the antibody library can be increased. The construction of synthesis library does not require immunity, and can also be applied to non-immunogenic targets. However, the obtained nanobodies may require improved affinity and stability, and require very large library scale (109-1015).
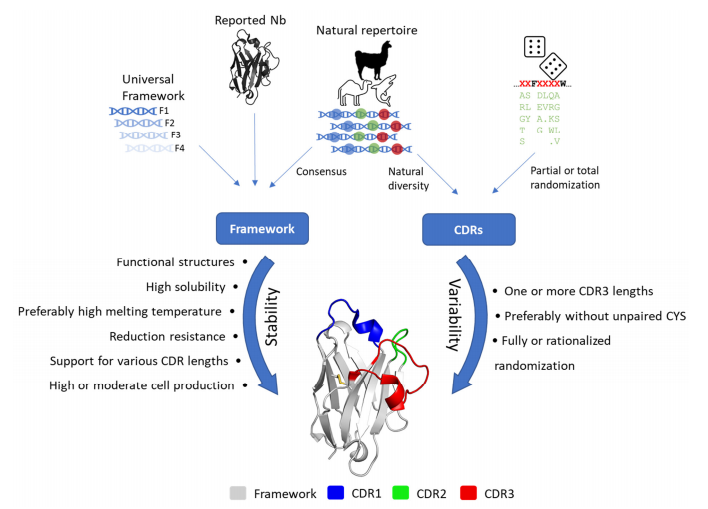
Figure 4. Strategy and desired characteristics for the generation of synthetic libraries.
(Ernest et al., 2022)
However, phage surface display technology is extremely dependent on antibody libraries with high library capacity and rich diversity, and phage manipulation is easy to contaminate, spread and difficult to control. Some laboratories also use other library display techniques, such as yeast display and ribosome display. The biggest drawback of these techniques is that there is not enough antibody diversity and a lot of antibody information is lost in the process of library preparation. Moreover, due to the steps of RNA extraction and reverse transcription, which are highly susceptible to contamination and degradation, it is difficult to ensure a HTP screening library capacity, so that the subsequent reverse transcription and PCR amplification are easily affected by the specificity and quantity of the template, resulting in screening failure or prolonged screening cycle.
Therefore, the technical path of obtaining nanobodies based on Single B cell cloning is a better choice. The Single B rapid screening platform is a short-term, simple, diverse, efficient and phage-free antibody screening method. It combines flow cell sorting and high-throughput on chip sequencing technologies to directly sort individual B cells and sequence their antibody-coding gene to obtain antibody sequence information. This method can cover all B cells targeting antigen specificity at one time, with good genetic diversity and high efficiency, and reduce the loss of comprehensive coverage of positive antibodies when constructing libraries. At the same time, it can avoid the problems of false positives and non-specific binding during the screening of monoclonal antibodies using phage libraries. Compared to the library building process, which requires a lot of time for panning, the Single B-cell screening process is often extremely fast, greatly reducing the project cycle.
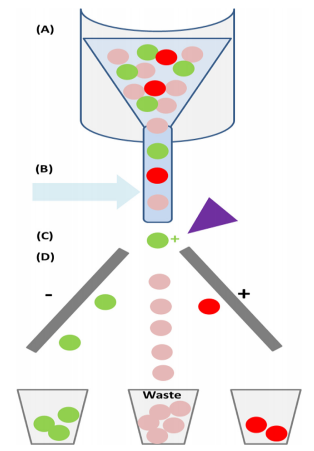
Figure 5. Fluorescence activated cell sorting (FACS).
(Fitzgerald & Leonard, 2017)
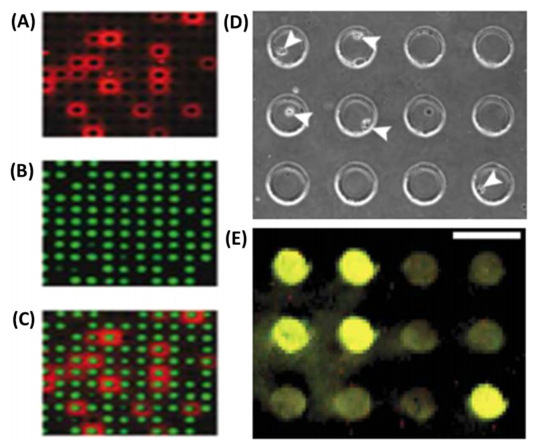
Figure 6. microengraved microwells.
(Fitzgerald & Leonard, 2017)
Comparison of Different Development Methods
| DetaiBio
SingleB® VHH | Phage Display Library |
|---|
| Synthesis Library | Natural Librar | Immune Library |
|---|
| Source of Antibody | Memory B cells and plasma cells | / | B cells, without immune animals | B cells, immune animals |
| Library Scale | - | 109-1015 | 109-1011 | 106-108 |
| Throughput | Very high | High | High | High |
| Screening Timeline | 1 day | 2 months | 2 months | 2 months |
| Antibody Diversity | High | Low | Low | Medium |
| Antibody Affinity | Very high | High | Low | High |
| Limitations | / | Depends on the bacteria in the correct transcription, translation, folding and assembly |
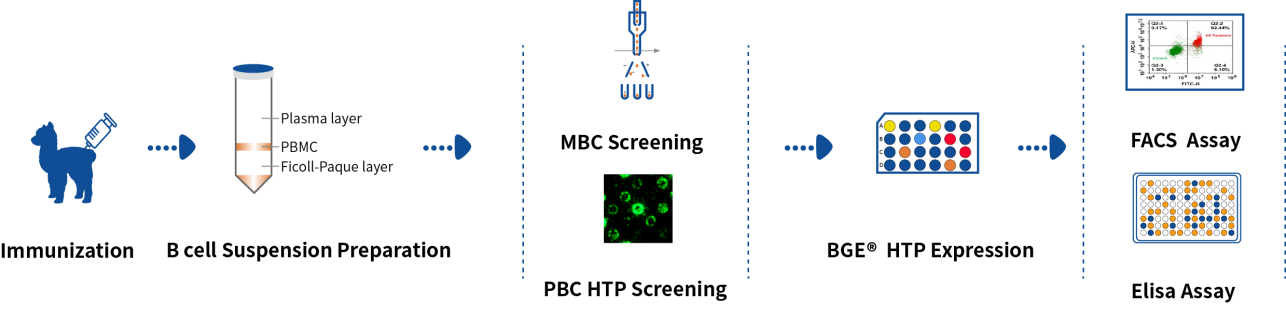
Alpaca SingleB® VHH Discovery Service
Based on the flow cytometry sorting platform and DeepLight® screening platform, DetaiBio provides alpaca VHH SingleB® rapid preparation service, from alpaca immunization, double screening of memory B cells and plasma cells, BGE® high-throughput expression, expression identification to purification, the whole process takes only 90 days, saving at least 15 days compared with immune display libraries.
Conclusion
Overall, the unique nature of nanobodies greatly broadens their application scope, especially for some targets that are difficult to be addressed by traditional techniques, which creates a huge market opportunity for nanobodies. Compared with library display, SingleB® technology has many advantages, such as fast speed, large delivery, good diversity and high affinity, which can avoid the lack of diversity due to limited library capacity and greatly reduce the preparation time of nanobodies.
Reference
1.Chen, X., Gentili, M., Hacohen, N. and Regev, A., 2021. A cell-free nanobody engineering platform rapidly generates SARS-CoV-2 neutralizing Nature communications, 12(1), p.5506.
2.Fitzgerald, V. and Leonard, P., 2017. single cell screening approaches for antibody discovery. methods, 116, pp. 34-42.
3.Lin, J., Gu, Y., Xu, Y., Yu, J., Tang, J., Wu, L., Zhou, Z., Chen, C., Liu, M., Chun, X. and Liu, H., 2020. Characterization and applications of nanobodies against Pseudomonas aeruginosa Exotoxin A selected from single alpaca B cells. Biotechnology & Biotechnological Equipment, 34(1), pp.1028-1037 .
4.Lyu, M., Shi, X., Liu, X., Liu, Y., Zhu, X., Liao, L., Zhao, H., Sun, N., Wang, S., Chen, L. and Fan, L., 2022. Generation and Screening of Antigen-Specific Nanobodies from Mammalian Cells Expressing the BCR Repertoire Library Using Droplet-Based Microfluidics. Analytical Chemistry, 94(22), pp.7970- 7980.
5.Salvador, J.P., Vilaplana, L. and Marco, M.P., 2019. Nanobody: outstanding features for diagnostic and therapeutic applications. Analytical and bioanalytical chemistry, 411, pp. 1703-1713.
6.Valdés-Tresanco, M.S., Molina-Zapata, A., Pose, A.G. and Moreno, E., 2022. Structural insights into the design of synthetic nanobody libraries. Molecules, 27(7), p.2198.










